
Introduction: Adjusted Predictions and Marginal Effects for Random Effects Models
Source:vignettes/introduction_randomeffects.Rmd
introduction_randomeffects.RmdThis vignette shows how to calculate adjusted predictions for mixed models. However, for mixed models, since random effects are involved, we can calculate conditional predictions and marginal predictions. We also have to distinguish between population-level and unit-level predictions.
But one thing at a time…
 Summary of most important points:
Summary of most important points:
-
Predictions can be made on the population-level or for each level of the grouping variable (unit-level). If unit-level predictions are requested, you need to set
type="random"and specify the grouping variable(s) in thetermsargument. -
Population-level predictions can be either conditional (predictions for a "typical" group) or marginal (average predictions across all groups). Set
margin="empirical"for marginal predictions. You'll notice differences in predictions especially for unequal group sizes at the random effects level. -
Prediction intervals, i.e. when
interval="prediction"also account for the uncertainty in the random effects.
Population-level predictions for mixed effects models
Mixed models are used to account for the dependency of observations within groups, e.g. repeated measurements within subjects, or students within schools. The dependency is modeled by random effects, i.e. mixed model at least have one grouping variable (or factor) as higher level unit.
At the lowest level, you have your fixed effects, i.e. your “independent variables” or “predictors”.
Adjusted predictions can now be calculated for specified values or levels of the focal terms, however, either for the full sample (population-level) or for each level of the grouping variable (unit-level). The latter is particularly useful when the grouping variable is of interest, e.g. when you want to compare the effect of a predictor between different groups.
To get population-level predictions, we set
type = "fixed" or type = "zero_inflation" (for
models with zero-inflation component).
Conditional and marginal effects/predictions
We start with the population-level predictions. Here you can either
calculate the conditional or the marginal effect (see
in detail also Heiss 2022). The conditional effect is the
effect of a predictor in an average or typical group, while the marginal
effect is the average effect of a predictor across all groups. E.g.
let’s say we have countries as grouping variable and
gdp (gross domestic product per capita) as predictor, then
the conditional and marginal effect would be:
conditional effect: effect of
gdpin an average or typical country. To get conditional predictions, we usepredict_response()orpredict_response(margin = "mean_mode").marginal effect: average effect of
gdpacross all countries. To get marginal (or average) predictions, we usepredict_response(margin = "empirical").
While the term “effect” referes to the strength of the relationship between a predictor and the response, “predictions” refer to the actual predicted values of the response. Thus, in the following, we will talk about conditional and marginal (or average) predictions (instead of effects).
In a balanced data set, where all groups have the same number of observations, the conditional and marginal predictions are often similar (maybe slightly different, depending on the non-focal predictors). However, in unbalanced data, the conditional and marginal predicted values can largely differ.
library(ggeffects)
library(lme4)
data(sleepstudy)
# balanced data set
m <- lmer(Reaction ~ Days + (1 + Days | Subject), data = sleepstudy)
# conditional predictions
predict_response(m, "Days [1,5,9]")
#> # Predicted values of Reaction
#>
#> Days | Predicted | 95% CI
#> ---------------------------------
#> 1 | 261.87 | 248.48, 275.27
#> 5 | 303.74 | 284.83, 322.65
#> 9 | 345.61 | 316.74, 374.48
#>
#> Adjusted for:
#> * Subject = 0 (population-level)
# average marginal predictions
predict_response(m, "Days [1,5,9]", margin = "empirical")
#> # Average predicted values of Reaction
#>
#> Days | Predicted | 95% CI
#> ---------------------------------
#> 1 | 261.87 | 248.48, 275.27
#> 5 | 303.74 | 284.83, 322.65
#> 9 | 345.61 | 316.74, 374.48
# create imbalanced data set
set.seed(123)
strapped <- sleepstudy[sample.int(nrow(sleepstudy), nrow(sleepstudy), replace = TRUE), ]
m <- lmer(Reaction ~ Days + (1 + Days | Subject), data = strapped)
# conditional predictions
predict_response(m, "Days [1,5,9]")
#> # Predicted values of Reaction
#>
#> Days | Predicted | 95% CI
#> ---------------------------------
#> 1 | 261.49 | 246.57, 276.40
#> 5 | 302.13 | 281.30, 322.97
#> 9 | 342.78 | 311.19, 374.37
#>
#> Adjusted for:
#> * Subject = 0 (population-level)
# average marginal predictions
predict_response(m, "Days [1,5,9]", margin = "empirical")
#> # Average predicted values of Reaction
#>
#> Days | Predicted | 95% CI
#> ---------------------------------
#> 1 | 259.04 | 244.13, 273.95
#> 5 | 300.01 | 279.17, 320.84
#> 9 | 340.97 | 309.37, 372.56Population-level predictions and the REML argument
The conditional predictions returned by
predict_response() for the default marginalization
(i.e. when margin = "mean_reference" or
"mean_mode") may differ…
- depending on the setting of the
REMLargument during model fitting; - and depending on whether factors are included in the model or not.
library(glmmTMB)
set.seed(123)
sleepstudy$x <- as.factor(sample(1:3, nrow(sleepstudy), replace = TRUE))
# REML is FALSE
m1 <- glmmTMB(Reaction ~ Days + x + (1 + Days | Subject), data = sleepstudy, REML = FALSE)
# REML is TRUE
m2 <- glmmTMB(Reaction ~ Days + x + (1 + Days | Subject), data = sleepstudy, REML = TRUE)
# predictions when REML is FALSE
predict_response(m1, "Days [1:3]")
#> # Predicted values of Reaction
#>
#> Days | Predicted | 95% CI
#> ---------------------------------
#> 1 | 260.22 | 245.82, 274.63
#> 2 | 270.69 | 255.77, 285.61
#> 3 | 281.16 | 265.19, 297.12
#>
#> Adjusted for:
#> * x = 1
#> * Subject = NA (population-level)
# predictions when REML is TRUE
predict_response(m2, "Days [1:3]")
#> # Predicted values of Reaction
#>
#> Days | Predicted | 95% CI
#> ---------------------------------
#> 1 | 254.63 | 246.25, 263.02
#> 2 | 265.07 | 257.33, 272.81
#> 3 | 275.50 | 268.22, 282.78
#>
#> Adjusted for:
#> * x = 1
#> * Subject = NA (population-level)Population-level predictions for zero-inflated mixed models
For zero-inflated mixed effects models, typically fitted with the
glmmTMB or GLMMadaptive packages,
predict_response() can return predicted values of the
response, for the different model components:
- Conditional predictions:
- population-level predictions, conditioned on the fixed effects
(conditional or “count” model) only (
type = "fixed") - population-level predictions, conditioned on the fixed effects
and zero-inflation component
(
type = "zero_inflated"), returning the expected (predicted) values of the response - the zero-inflation probabilities
(
type = "zi_prob")
- population-level predictions, conditioned on the fixed effects
(conditional or “count” model) only (
- Marginal predictions:
-
type = "simulate"can be used to obtain marginal predictions, averaged across all random effects groups and non-focal terms - marginal predictions using
margin = "empirical"are also averaged across all random effects groups and non-focal terms. The major difference totype = "simulate"is thatmargin = "empirical"also returns counterfactual predictions.
-
For predict_response(margin = "empirical"), valid values
for type are usually those based on the model’s
predict() method. For models of class glmmTMB,
these are "response", "link",
"conditional", "zprob", "zlink",
or "disp". However, for zero-inflated models,
type = "fixed" and type = "zero_inflated" can
be used as aliases (instead of "conditional" or
"response").
Conditional predictions for the count model
First, we show examples for conditional predictions, which is the
default marginalization method in predict_response().
library(glmmTMB)
data(Salamanders)
m <- glmmTMB(
count ~ spp + mined + (1 | site),
ziformula = ~ spp + mined,
family = poisson(),
data = Salamanders
)Similar to mixed models without zero-inflation component,
type = "fixed" returns predictions on the population-level,
but for the conditional (“count”) model only.
predict_response(m, "spp")
#> # Predicted (conditional) counts of count
#>
#> spp | Predicted | 95% CI
#> ------------------------------
#> GP | 0.73 | 0.42, 1.28
#> PR | 0.42 | 0.20, 0.87
#> DM | 0.94 | 0.56, 1.58
#> EC-A | 0.60 | 0.33, 1.10
#> EC-L | 1.42 | 0.85, 2.37
#> DES-L | 1.34 | 0.79, 2.26
#> DF | 0.78 | 0.46, 1.31
#>
#> Adjusted for:
#> * mined = yes
#> * site = NA (population-level)Conditional predictions for the full model
For type = "zero_inflated", results the expected values
of the response, mu*(1-p). Since the zero inflation and the
conditional model are working in “opposite directions”, a higher
expected value for the zero inflation means a lower response, but a
higher value for the conditional (“count”) model means a higher
response. While it is possible to calculate predicted values with
predict(..., type = "response"), standard errors and
confidence intervals can not be derived directly from the
predict()-function. Thus, confidence intervals for
type = "zero_inflated" are based on quantiles of simulated
draws from a multivariate normal distribution (see also Brooks et
al. 2017, pp.391-392 for details).
predict_response(m, "spp", type = "zero_inflated")
#> # Expected counts of count
#>
#> spp | Predicted | 95% CI
#> ------------------------------
#> GP | 0.20 | 0.02, 0.38
#> PR | 0.03 | 0.00, 0.06
#> DM | 0.32 | 0.08, 0.56
#> EC-A | 0.07 | 0.00, 0.13
#> EC-L | 0.42 | 0.12, 0.73
#> DES-L | 0.49 | 0.13, 0.85
#> DF | 0.30 | 0.09, 0.51
#>
#> Adjusted for:
#> * mined = yes
#> * site = NA (population-level)Marginal predictions for the full model (simulated draws)
In the above examples, we get the conditional, not the marginal
predictions. E.g., predictions are conditioned on mined
when it is set to "yes", and predictions refer to a
typical (random effects) group. However, it is possible to
obtain predicted values by simulating from the model, where predictions
are based on simulate() (see Brooks et al. 2017,
pp.392-393 for details). This will return expected values of the
response (marginal predictions), averaged across all random
effects groups and non-focal terms. To achieve this, use
type = "simulate". Note that this prediction-type usually
returns larger intervals, because it accounts for all model
uncertainties.
predict_response(m, "spp", type = "simulate")
#> # Expected counts of count
#>
#> spp | Predicted | 95% CI
#> ------------------------------
#> GP | 1.10 | 0.00, 4.31
#> PR | 0.30 | 0.00, 2.23
#> DM | 1.54 | 0.00, 5.49
#> EC-A | 0.55 | 0.00, 3.11
#> EC-L | 2.20 | 0.00, 7.35
#> DES-L | 2.26 | 0.00, 7.14
#> DF | 1.35 | 0.00, 4.72Marginal predictions for the full model (average predictions)
In a similar fashion, you can obtain average marginal predictions for
zero-inflated mixed models with margin = "empirical". The
returned values are most comparable to
predict_response(type = "simulate"), because
margin = "empirical" also returns expected values of the
response, averaged across all random effects groups and all non-focal
terms. The next example shows the average marginal predicted values of
spp on the response across all sites, taking
the zero-inflation component into account
(i.e. type = "zero_inflated").
predict_response(m, "spp", type = "zero_inflated", margin = "empirical")
#> # Average expected counts of count
#>
#> spp | Predicted | 95% CI
#> ------------------------------
#> GP | 1.08 | 0.76, 1.41
#> PR | 0.30 | 0.13, 0.46
#> DM | 1.52 | 1.11, 1.94
#> EC-A | 0.54 | 0.31, 0.78
#> EC-L | 2.17 | 1.60, 2.74
#> DES-L | 2.24 | 1.69, 2.79
#> DF | 1.32 | 0.96, 1.68Bias-correction for non-Gaussian models
For non-Gaussian models, predicted values are back-transformed to the
response scale. However, back-transforming the population-level
predictions (in mixed models, when type = "fixed")
ignores the effect of the variation around the population mean, hence,
the result on the original data scale is biased due to Jensen’s
inequality. In this case, it can be appropriate to apply a
bias-correction. This is done by setting
bias_correction = TRUE. By default, insight::get_variance_residual()
is used to extract the residual variance, which is used to calculate the
amount of correction. Optionally, you can provide your own estimates of
uncertainty, e.g. based on insight::get_variance_random(),
using the sigma argument. ggeffects will warn
users once per session whenever bias-correction can be appropriate.
# no bias-correction
predict_response(m, "spp")
#> You are calculating adjusted predictions on the population-level (i.e. `type = "fixed"`) for a *generalized* linear mixed model.
#> This may produce biased estimates due to Jensen's inequality. Consider setting `bias_correction = TRUE` to correct for this bias.
#> See also the documentation of the `bias_correction` argument.
#> # Predicted (conditional) counts of count
#>
#> spp | Predicted | 95% CI
#> ------------------------------
#> GP | 0.73 | 0.42, 1.28
#> PR | 0.42 | 0.20, 0.87
#> DM | 0.94 | 0.56, 1.58
#> EC-A | 0.60 | 0.33, 1.10
#> EC-L | 1.42 | 0.85, 2.37
#> DES-L | 1.34 | 0.79, 2.26
#> DF | 0.78 | 0.46, 1.31
#>
#> Adjusted for:
#> * mined = yes
#> * site = NA (population-level)
# bias-correction
predict_response(m, "spp", bias_correction = TRUE)
#> # Predicted (conditional) counts of count
#>
#> spp | Predicted | 95% CI
#> ------------------------------
#> GP | 0.97 | 0.55, 1.69
#> PR | 0.55 | 0.27, 1.15
#> DM | 1.24 | 0.73, 2.09
#> EC-A | 0.79 | 0.43, 1.45
#> EC-L | 1.87 | 1.12, 3.12
#> DES-L | 1.76 | 1.04, 2.98
#> DF | 1.02 | 0.60, 1.74
#>
#> Adjusted for:
#> * mined = yes
#> * site = NA (population-level)
# bias-correction, using user-defined sigma-value
predict_response(m, "spp", bias_correction = TRUE, sigma = insight::get_sigma(m))
#> # Predicted (conditional) counts of count
#>
#> spp | Predicted | 95% CI
#> ------------------------------
#> GP | 1.10 | 0.63, 1.92
#> PR | 0.63 | 0.30, 1.30
#> DM | 1.41 | 0.83, 2.38
#> EC-A | 0.90 | 0.49, 1.65
#> EC-L | 2.13 | 1.27, 3.55
#> DES-L | 2.01 | 1.19, 3.39
#> DF | 1.16 | 0.69, 1.97
#>
#> Adjusted for:
#> * mined = yes
#> * site = NA (population-level)Unit-level predictions (predictions for each level of random effects)
Adjusted predictions can also be calculated for each group level
(unit-level) in mixed models. Simply add the name of the related random
effects term to the terms-argument, and set
type = "random". For
predict_response(margin = "empirical"), you don’t need to
set type = "random".
In the following example, we fit a linear mixed model and first simply plot the adjusted predictions at the population-level.
library(sjlabelled)
data(efc)
efc$e15relat <- as_label(efc$e15relat)
m <- lmer(neg_c_7 ~ c12hour + c160age + c161sex + (1 | e15relat), data = efc)
me <- predict_response(m, terms = "c12hour")
plot(me)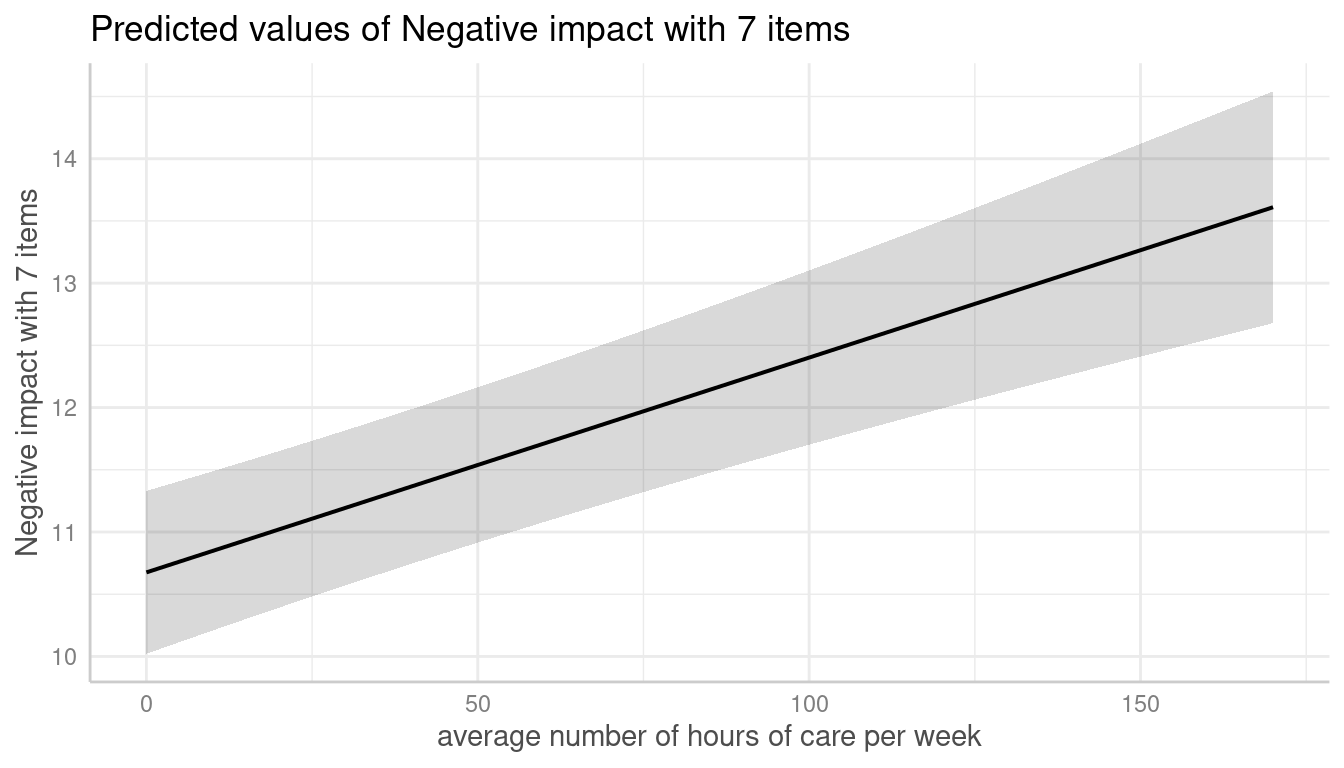
To compute adjusted predictions for each grouping level (unit-level),
add the related random effects term to the terms-argument.
In this case, predictions are calculated for each level of the specified
random effects term.
me <- predict_response(m, terms = c("c12hour", "e15relat"), type = "random")
plot(me, show_ci = FALSE)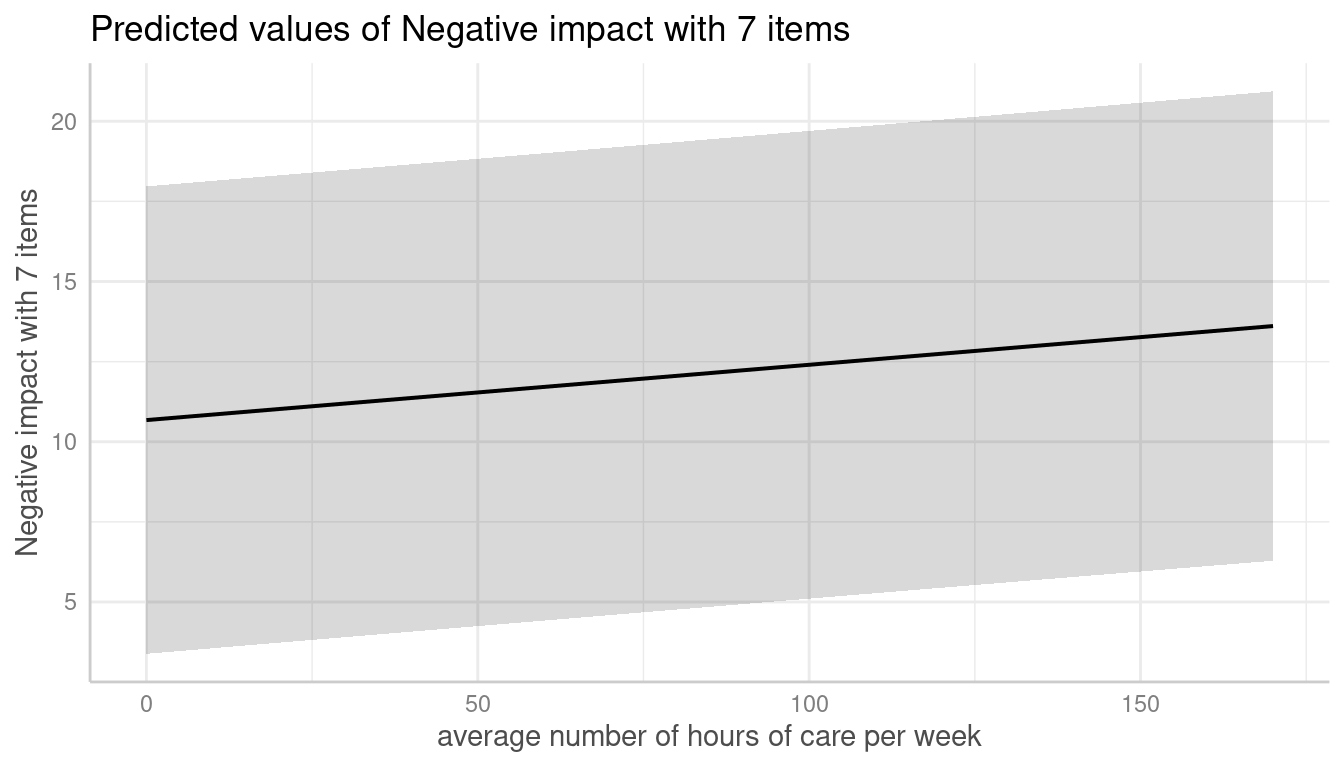
Since average marginal predictions already consider random effects by
averaging over the groups, the type-argument is not needed
when margin = "empirical" is set.
me <- predict_response(m, terms = c("c12hour", "e15relat"), margin = "empirical")
plot(me, show_ci = FALSE)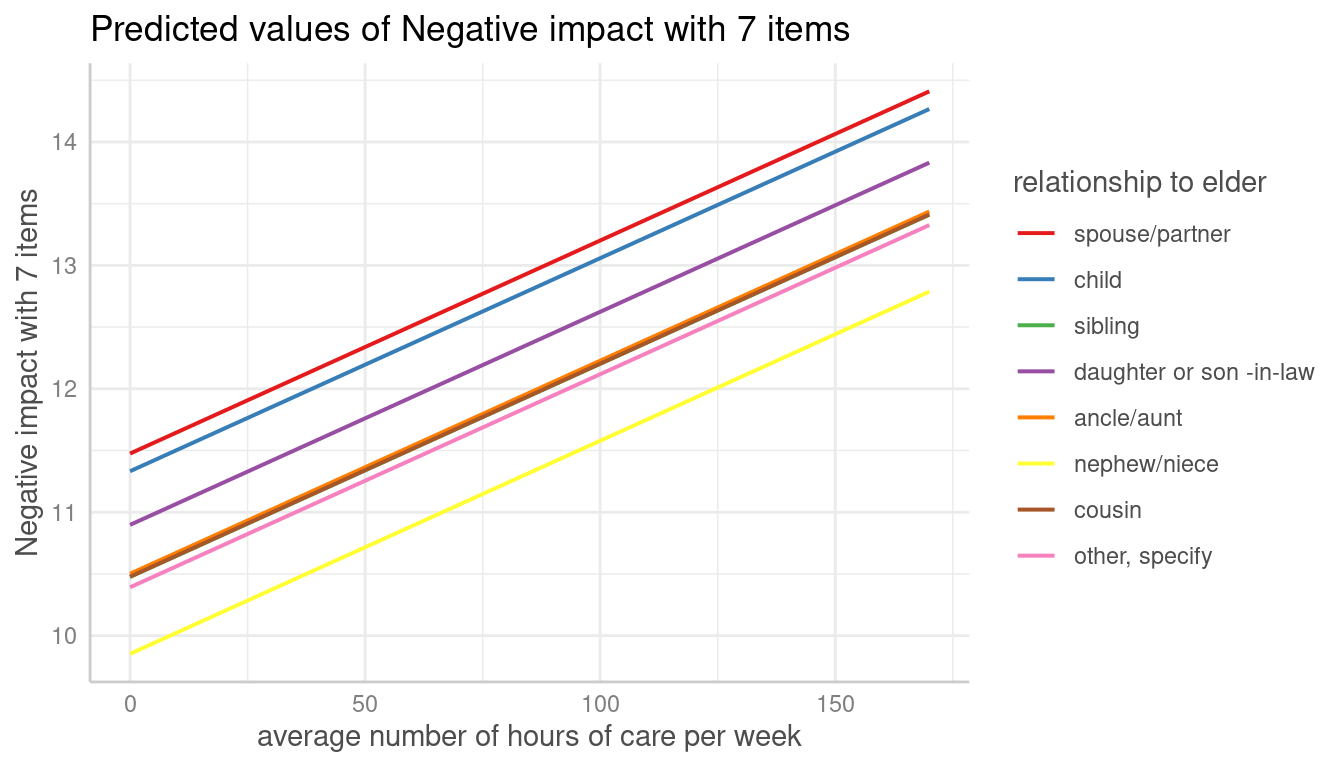
Adjusted predictions can also be calculated for specific unit-levels
only. Add the related values into brackets after the variable name in
the terms-argument.
me <- predict_response(m, terms = c("c12hour", "e15relat [child,sibling]"), type = "random")
plot(me, show_ci = FALSE)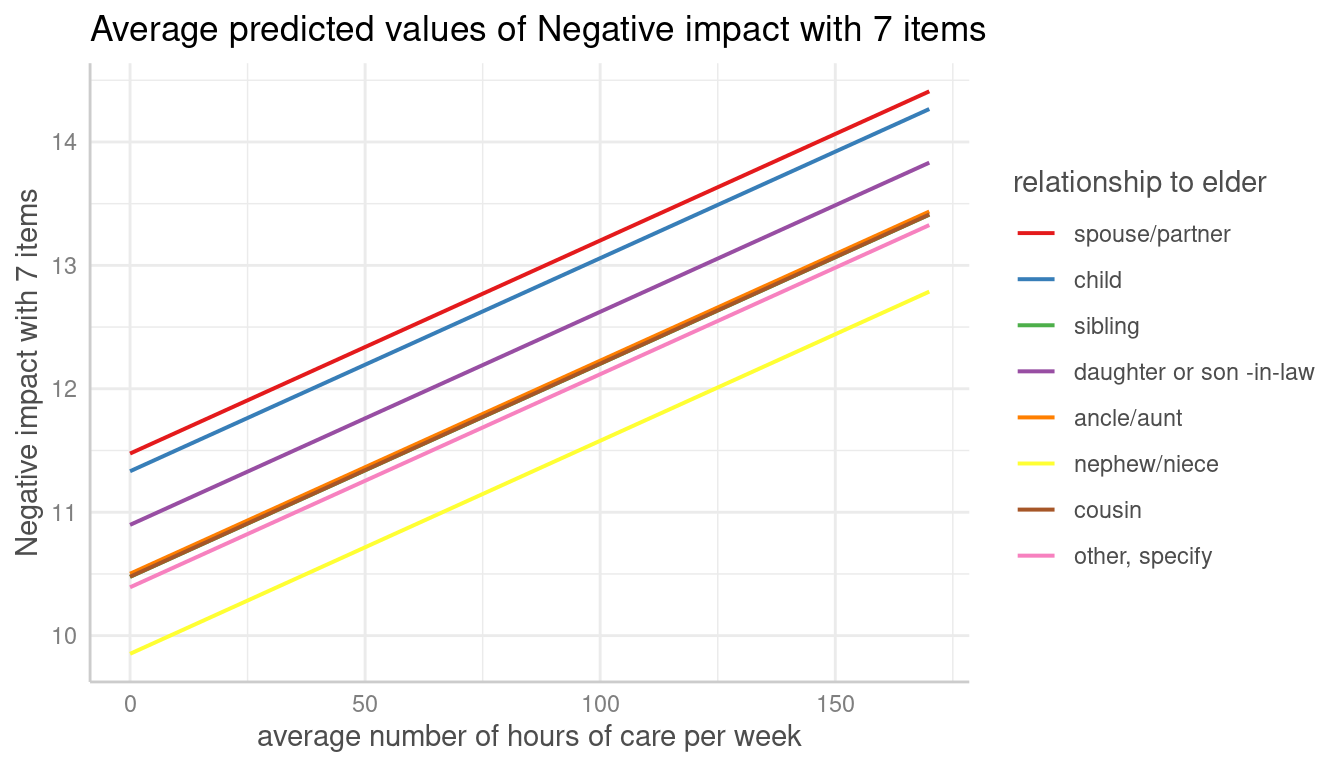
The complex plot in this scenario would be a term
(c12hour) at certain values of two other terms
(c161sex, c160age) for specific unit-levels of
random effects (e15relat), so we have four variables in the
terms-argument.
me <- predict_response(
m,
terms = c("c12hour", "c161sex", "c160age", "e15relat [child,sibling]"),
type = "random"
)
plot(me)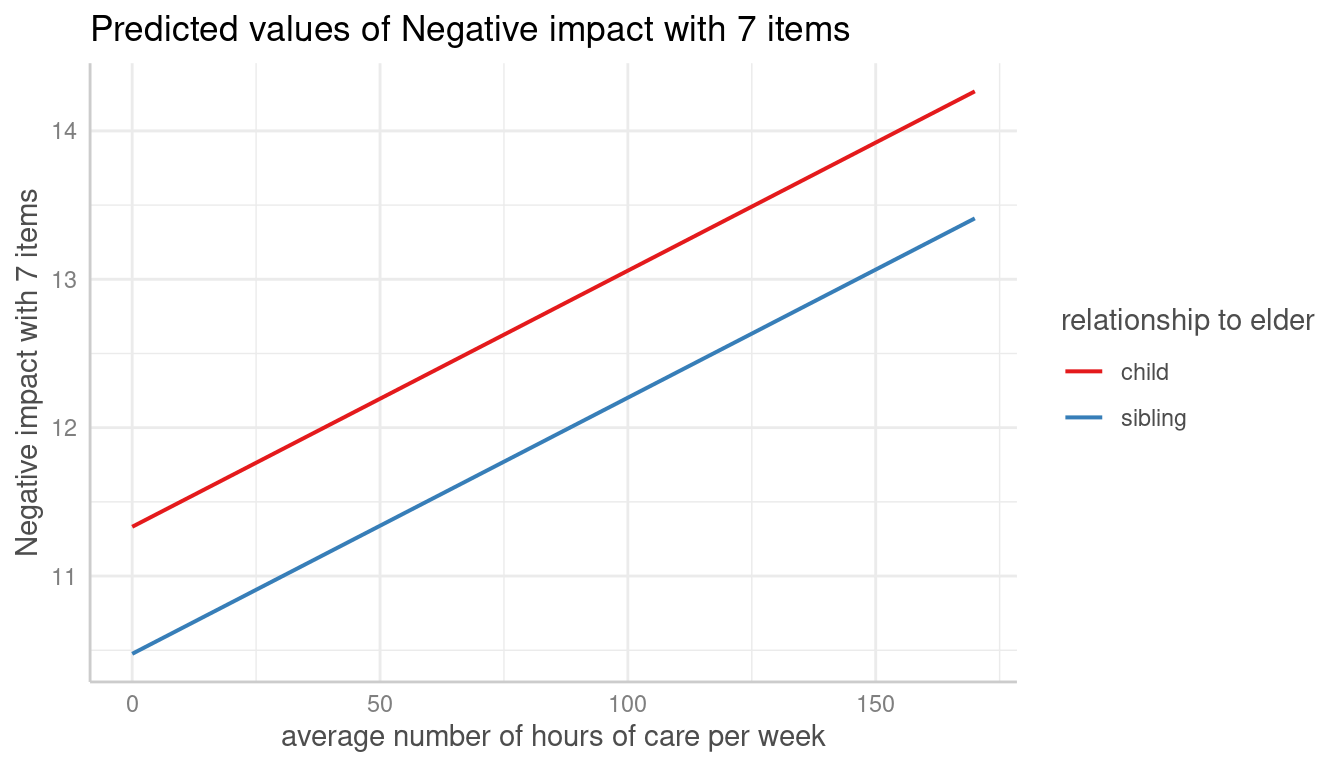
If the group factor has too many levels, you can also take a random
sample of all possible levels and plot the adjusted predictions for this
subsample of unit-levels. To do this, use
term = "<groupfactor> [sample=n]".
set.seed(123)
m <- lmer(Reaction ~ Days + (1 + Days | Subject), data = sleepstudy)
me <- predict_response(m, terms = c("Days", "Subject [sample=7]"), type = "random")
plot(me)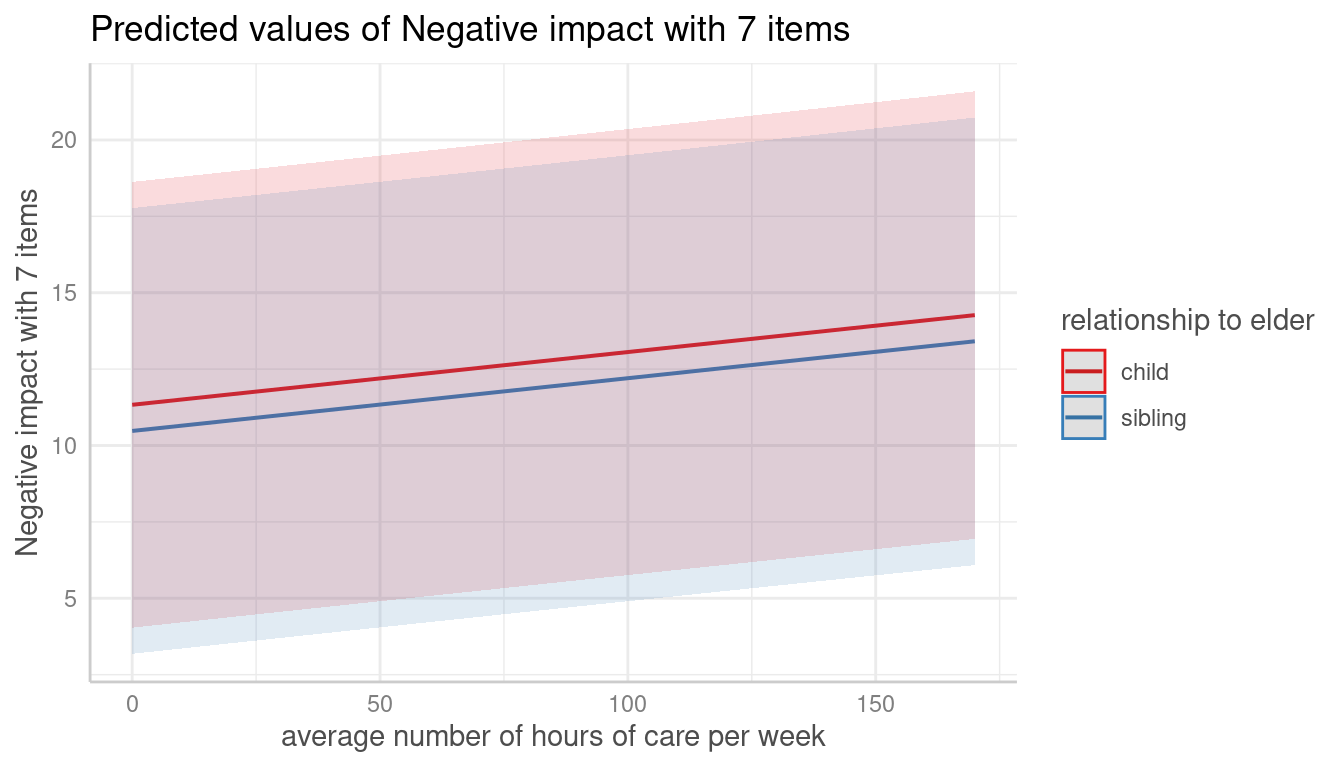
You can also add the observed data points for each group using
show_data = TRUE.
plot(me, show_data = TRUE, show_ci = FALSE)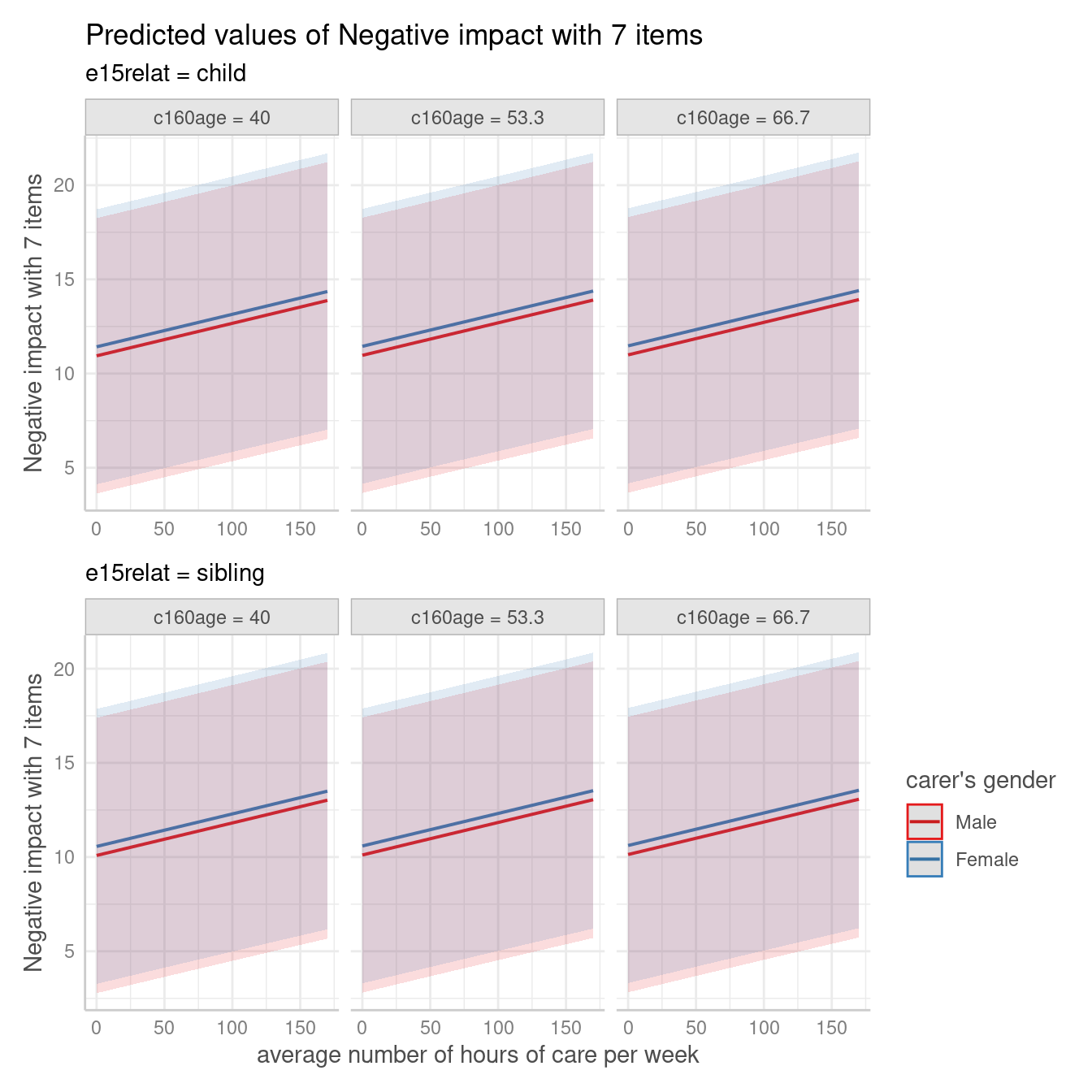
Population-level predictions for gam and
glmer models
The output of predict_response() indicates that the
grouping variable of the random effects is set to “population level”
(adjustment), e.g. in case of lme4, following is printed:
Adjusted for: * Subject = 0 (population-level)
A comparable model fitted with mgcv::gam() would print a
different message:
Adjusted for: * Subject = 308
The reason is because the correctly printed information about
adjustment for random effects is based on
insight::find_random(), which returns NULL for
gams with random effects defined via
s(..., bs = "re"). However, predictions are still correct,
when population-level predictions are requested. Here’s an example:
data("sleepstudy", package = "lme4")
# mixed model with lme4
m_lmer <- lme4::lmer(Reaction ~ poly(Days, 2) + (1 | Subject),
data = sleepstudy
)
# equivalent model, random effects are defined via s(..., bs = "re")
m_gam <- mgcv::gam(Reaction ~ poly(Days, 2) + s(Subject, bs = "re"),
family = gaussian(), data = sleepstudy, method = "ML"
)
# predictions are identical
predict_response(m_gam, terms = "Days", exclude = "s(Subject)", newdata.guaranteed = TRUE)
#> # Predicted values of Reaction
#>
#> Days | Predicted | 95% CI
#> ---------------------------------
#> 0 | 255.45 | 235.12, 275.78
#> 1 | 263.22 | 244.71, 281.73
#> 2 | 271.67 | 253.70, 289.63
#> 3 | 280.78 | 262.75, 298.82
#> 5 | 301.05 | 282.84, 319.25
#> 6 | 312.19 | 294.15, 330.22
#> 7 | 324.00 | 306.03, 341.97
#> 9 | 349.65 | 329.33, 369.98
#>
#> Adjusted for:
#> * Subject = 308
predict_response(m_lmer, terms = "Days")
#> # Predicted values of Reaction
#>
#> Days | Predicted | 95% CI
#> ---------------------------------
#> 0 | 255.45 | 234.79, 276.10
#> 1 | 263.22 | 244.35, 282.09
#> 2 | 271.67 | 253.33, 290.00
#> 3 | 280.78 | 262.38, 299.19
#> 5 | 301.05 | 282.48, 319.61
#> 6 | 312.19 | 293.78, 330.59
#> 7 | 324.00 | 305.66, 342.34
#> 9 | 349.65 | 329.00, 370.31
#>
#> Adjusted for:
#> * Subject = 0 (population-level)References
Brooks ME, Kristensen K, Benthem KJ van, Magnusson A, Berg CW, Nielsen A, et al. glmmTMB Balances Speed and Flexibility Among Packages for Zero-inflated Generalized Linear Mixed Modeling. The R Journal. 2017;9: 378–400.
Heiss, A. (2022, November 29). Marginal and conditional effects for GLMMs with {marginaleffects}. Andrew Heiss’s Blog. (doi: 10.59350/xwnfm-x1827)
Johnson PC. 2014. Extension of Nakagawa & Schielzeth’s R2GLMM to random slopes models. Methods Ecol Evol, 5: 944-946. (doi: 10.1111/2041-210X.12225)