
Technical Details: Difference Between Marginalization Methods: The `margin` Argument
Source:vignettes/technical_differencepredictemmeans.Rmd
technical_differencepredictemmeans.Rmdpredict_response() computes marginal means or predicted
values for all possible levels or values from specified model’s
predictors (focal terms). These effects are “marginalized” (or
“averaged”) over the values or levels of remaining predictors (the
non-focal terms). The margin argument
specifies the method of marginalization. The following methods are
available:
"mean_reference": non-focal predictors are set to their mean (numeric variables), reference level (factors), or “most common” value (mode) in case of character vectors. Technically, a data grid is constructed, roughly comparable toexpand.grid()on all unique combinations ofmodel.frame(model)[, terms]. This data grid (seedata_grid()) is used for thenewdataargument ofpredict(). All remaining covariates not specified intermsare held constant: Numeric values are set to the mean, integer values are set to their median, factors are set to their reference level and character vectors to their mode (most common element)."mean_mode": non-focal predictors are set to their mean (numeric variables) or mode (factors, or “most common” value in case of character vectors)."marginalmeans": non-focal predictors are set to their mean (numeric variables) or marginalized over the levels or “values” for factors and character vectors. Marginalizing over the factor levels of non-focal terms computes a kind of “weighted average” for the values at which these terms are hold constant."empirical"(or"counterfactual"): non-focal predictors are marginalized over the observations in your sample, for counterfactual predictions. Technically, predicted values for each observation in the data are calculated multiple times (the data is duplicated once for all unique values of the focal terms), each time fixing one unique value or level of the focal terms and then takes the average of these predicted values (aggregated/grouped by the focal terms).
That means:
For models without categorical predictors, results are usually identical, no matter which
marginoption is selected (except some slight differences in the associated confidence intervals, which are, however, negligible).When all categorical predictors are specified in
termsand further (non-focal) terms are only numeric, results are usually identical, as well.
In the general introduction, the
margin argument is discussed more in detail, providing
hints on when to use which method. Here, we will provide a more
technical explanation of the differences between the methods.
library(ggeffects)
data(efc, package = "ggeffects")
fit <- lm(barthtot ~ c12hour + neg_c_7, data = efc)
# we add margin = "mean_reference" to show that it is the default
predict_response(fit, "c12hour [0,50,100,150]", margin = "mean_reference")
#> # Predicted values of Total score BARTHEL INDEX
#>
#> c12hour | Predicted | 95% CI
#> ----------------------------------
#> 0 | 75.07 | 72.96, 77.19
#> 50 | 62.78 | 61.16, 64.40
#> 100 | 50.48 | 48.00, 52.97
#> 150 | 38.19 | 34.30, 42.08
#>
#> Adjusted for:
#> * neg_c_7 = 11.83
predict_response(fit, "c12hour [0,50,100,150]", margin = "marginalmeans")
#> # Predicted values of Total score BARTHEL INDEX
#>
#> c12hour | Predicted | 95% CI
#> ----------------------------------
#> 0 | 75.07 | 72.96, 77.19
#> 50 | 62.78 | 61.16, 64.40
#> 100 | 50.48 | 48.00, 52.97
#> 150 | 38.19 | 34.30, 42.08
#>
#> Adjusted for:
#> * neg_c_7 = 11.83As can be seen, the continuous predictor neg_c_7 is held
constant at its mean value, 11.83. For categorical predictors,
margin = "mean_reference" (the default, and thus not
specified in the above example) and
margin = "marginalmeans" behave differently. While
"mean_reference" uses the reference level of each
categorical predictor to hold it constant, "marginalmeans"
averages over the proportions of the categories of factors.
library(datawizard)
data(efc, package = "ggeffects")
efc$e42dep <- to_factor(efc$e42dep)
# we add categorical predictors to our model
fit <- lm(barthtot ~ c12hour + neg_c_7 + e42dep, data = efc)
predict_response(fit, "c12hour [0,50,100,150]", margin = "mean_reference")
#> # Predicted values of Total score BARTHEL INDEX
#>
#> c12hour | Predicted | 95% CI
#> ----------------------------------
#> 0 | 92.74 | 88.48, 97.01
#> 50 | 89.18 | 84.82, 93.54
#> 100 | 85.61 | 80.79, 90.42
#> 150 | 82.04 | 76.49, 87.59
#>
#> Adjusted for:
#> * neg_c_7 = 11.83
#> * e42dep = independent
predict_response(fit, "c12hour [0,50,100,150]", margin = "marginalmeans")
#> # Predicted values of Total score BARTHEL INDEX
#>
#> c12hour | Predicted | 95% CI
#> ----------------------------------
#> 0 | 73.51 | 71.85, 75.18
#> 50 | 69.95 | 68.51, 71.38
#> 100 | 66.38 | 64.19, 68.56
#> 150 | 62.81 | 59.51, 66.10
#>
#> Adjusted for:
#> * neg_c_7 = 11.83In this case, one would obtain the same results for
"mean_reference" and "marginalmeans" again, if
condition is used to define specific levels at which
variables, in our case the factor e42dep, should be held
constant.
predict_response(fit, "c12hour [0,50,100,150]", margin = "mean_reference")
#> # Predicted values of Total score BARTHEL INDEX
#>
#> c12hour | Predicted | 95% CI
#> ----------------------------------
#> 0 | 92.74 | 88.48, 97.01
#> 50 | 89.18 | 84.82, 93.54
#> 100 | 85.61 | 80.79, 90.42
#> 150 | 82.04 | 76.49, 87.59
#>
#> Adjusted for:
#> * neg_c_7 = 11.83
#> * e42dep = independent
predict_response(
fit,
"c12hour [0,50,100,150]",
margin = "marginalmeans",
condition = c(e42dep = "independent")
)
#> # Predicted values of Total score BARTHEL INDEX
#>
#> c12hour | Predicted | 95% CI
#> ----------------------------------
#> 0 | 92.74 | 88.48, 97.01
#> 50 | 89.18 | 84.82, 93.54
#> 100 | 85.61 | 80.79, 90.42
#> 150 | 82.04 | 76.49, 87.59
#>
#> Adjusted for:
#> * neg_c_7 = 11.83Another option is to use
predict_response(margin = "empirical") to compute
“counterfactual” adjusted predictions. This function is a wrapper for
the avg_predictions()-method from the
marginaleffects-package. The major difference to
margin = "marginalmeans" is that estimated marginal means,
as computed by "marginalmeans", are a special case of
predictions, made on a perfectly balanced grid of categorical
predictors, with numeric predictors held at their means, and
marginalized with respect to some focal variables.
predict_response(margin = "empirical"), in turn,
calculates predicted values for each observation in the data multiple
times, each time fixing the unique values or levels of the focal terms
to one specific value and then takes the average of these predicted
values (aggregated/grouped by the focal terms) - or in other words: the
whole dataset is duplicated once for every unique value of the focal
terms, makes predictions for each observation of the new dataset and
take the average of all predictions (grouped by focal terms). This is
also called “counterfactual” predictions.
predict_response(fit, "c12hour", margin = "empirical")
#> # Average predicted values of Total score BARTHEL INDEX
#>
#> c12hour | Predicted | 95% CI
#> ----------------------------------
#> 0 | 67.73 | 66.15, 69.31
#> 20 | 66.30 | 65.03, 67.57
#> 45 | 64.52 | 63.38, 65.66
#> 65 | 63.09 | 61.81, 64.37
#> 85 | 61.66 | 60.07, 63.25
#> 105 | 60.23 | 58.24, 62.23
#> 125 | 58.81 | 56.37, 61.25
#> 170 | 55.59 | 52.08, 59.11To explain how margin = "empirical" works, let’s look at
following example, where we compute the average predicted values and the
estimated marginal means manually. The confidence intervals for the
manually calculated means differ from those of
predict_response(), however, the predicted and manually
calculated mean values are identical.
data(iris)
set.seed(123)
iris$x <- as.factor(sample(1:4, nrow(iris), replace = TRUE, prob = c(0.1, 0.2, 0.3, 0.4)))
m <- lm(Sepal.Width ~ Species + x, data = iris)
# average predicted values
predict_response(m, "Species", margin = "empirical")
#> # Average predicted values of Sepal.Width
#>
#> Species | Predicted | 95% CI
#> -----------------------------------
#> setosa | 3.43 | 3.34, 3.52
#> versicolor | 2.77 | 2.67, 2.86
#> virginica | 2.97 | 2.88, 3.07
# replicate the dataset for each level of "Species", i.e. 3 times
d <- do.call(rbind, replicate(3, iris, simplify = FALSE))
# for each data set, we set our focal term to one of the three levels
d$Species <- as.factor(rep(levels(iris$Species), each = 150))
# we calculate predicted values for each "dataset", i.e. we predict our outcome
# for observations, for all levels of "Species"
d$predicted <- predict(m, newdata = d)
# now we compute the average predicted values by levels of "Species", where
# non-focal terms are weighted proportional to their occurence in the data
datawizard::means_by_group(d, "predicted", "Species")
#> # Mean of predicted by Species
#>
#> Category | Mean | N | SD | 95% CI | p
#> ------------------------------------------------------
#> setosa | 3.43 | 150 | 0.06 | [3.42, 3.44] | < .001
#> versicolor | 2.77 | 150 | 0.06 | [2.76, 2.78] | < .001
#> virginica | 2.97 | 150 | 0.06 | [2.96, 2.98] | < .001
#> Total | 3.06 | 450 | 0.28 | |
#>
#> Anova: R2=0.951; adj.R2=0.951; F=4320.998; p<.001
# estimated marginal means, in turn, differ from the above, because they are
# averaged across balanced reference grids for all focal terms, thereby non-focal
# are hold constant at an (equally) "weighted average".
# estimated marginal means, from `ggemmeans()`
predict_response(m, "Species", margin = "marginalmeans")
#> # Predicted values of Sepal.Width
#>
#> Species | Predicted | 95% CI
#> -----------------------------------
#> setosa | 3.45 | 3.35, 3.54
#> versicolor | 2.78 | 2.68, 2.89
#> virginica | 2.99 | 2.89, 3.09
d <- rbind(
data_grid(m, "Species", condition = c(x = "1")),
data_grid(m, "Species", condition = c(x = "2")),
data_grid(m, "Species", condition = c(x = "3")),
data_grid(m, "Species", condition = c(x = "4"))
)
d$predicted <- predict(m, newdata = d)
# means calculated manually
datawizard::means_by_group(d, "predicted", "Species")
#> # Mean of predicted by Species
#>
#> Category | Mean | N | SD | 95% CI | p
#> -----------------------------------------------------
#> setosa | 3.45 | 4 | 0.07 | [3.36, 3.53] | < .001
#> versicolor | 2.78 | 4 | 0.07 | [2.70, 2.87] | < .001
#> virginica | 2.99 | 4 | 0.07 | [2.91, 3.07] | 0.019
#> Total | 3.07 | 12 | 0.30 | |
#>
#> Anova: R2=0.952; adj.R2=0.941; F=88.566; p<.001But when should I use which margin
option?
When you are interested in the strength of association, it usually
doesn’t matter. as you can see in the plots below. The slope of our
focal term, c12hour, is the same for all four plots:
library(see)
predicted_1 <- predict_response(fit, terms = "c12hour")
predicted_2 <- predict_response(fit, terms = "c12hour", margin = "marginalmeans")
predicted_3 <- predict_response(fit, terms = "c12hour", margin = "marginalmeans", condition = c(e42dep = "independent"))
predicted_4 <- predict_response(fit, terms = "c12hour", margin = "empirical")
p1 <- plot(predicted_1, show_ci = FALSE, show_title = FALSE, show_x_title = FALSE, show_y_title = FALSE)
p2 <- plot(predicted_2, show_ci = FALSE, show_title = FALSE, show_x_title = FALSE, show_y_title = FALSE)
p3 <- plot(predicted_3, show_ci = FALSE, show_title = FALSE, show_x_title = FALSE, show_y_title = FALSE)
p4 <- plot(predicted_4, show_ci = FALSE, show_title = FALSE, show_x_title = FALSE, show_y_title = FALSE)
plots(p1, p2, p3, p4, n_rows = 2)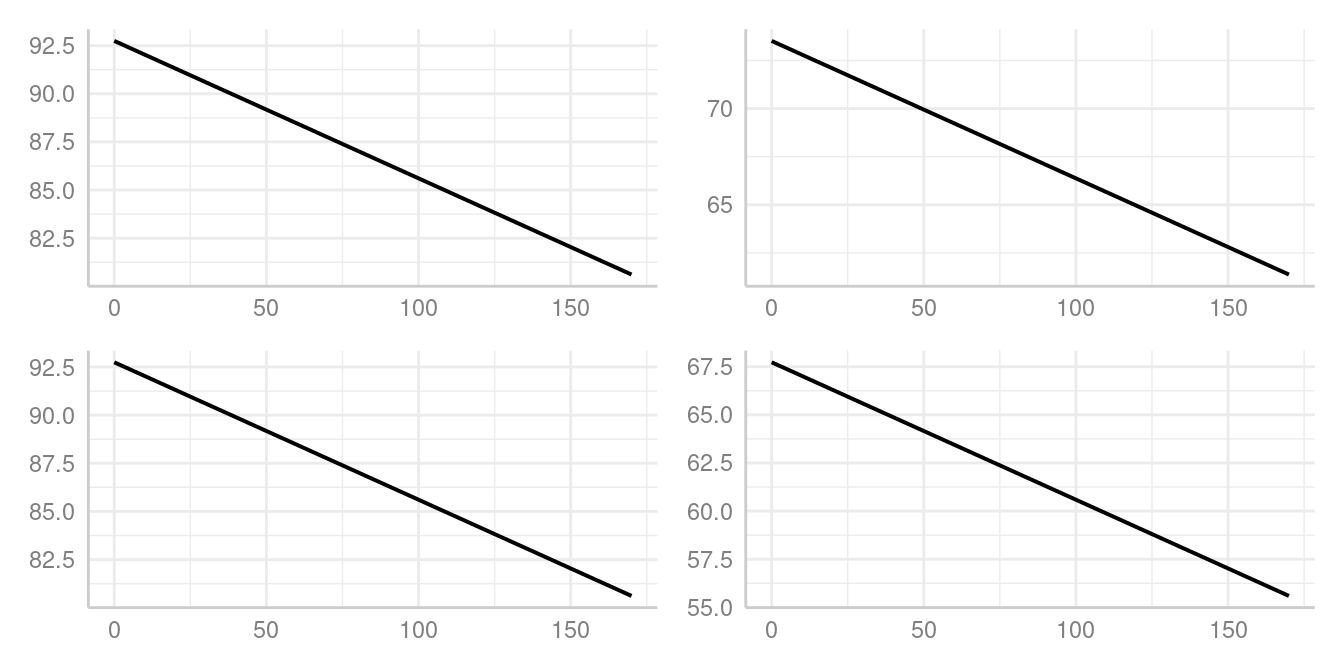
However, the predicted outcome varies. The general introduction discusses the
margin argument more in detail, but a few hints on when to
use which method are following:
Predictions based on
"mean_reference"and"mean_mode"represent a rather “theoretical” view on your data, which does not necessarily exactly reflect the characteristics of your sample. It helps answer the question, “What is the predicted value of the response at meaningful values or levels of my focal terms for a ‘typical’ observation in my data?”, where ‘typical’ refers to certain characteristics of the remaining predictors."marginalmeans"comes closer to the sample, because it takes all possible values and levels of your non-focal predictors into account. It would answer thr question, “What is the predicted value of the response at meaningful values or levels of my focal terms for an ‘average’ observation in my data?”. It refers to randomly picking a subject of your sample and the result you get on average."empirical"is probably the most “realistic” approach, insofar as the results can also be transferred to other contexts. It answers the question, “What is the predicted value of the response at meaningful values or levels of my focal terms for the ‘average’ observation in the population?”. It does not only refer to the actual data in your sample, but also “what would be if” we had more data, or if we had data from a different population. This is where “counterfactual” refers to.
What is the most apparent difference from
margin = "empirical" to the other options?
The most apparent difference from margin = "empirical"
compared to the other methods occurs when you have categorical
co-variates (non-focal terms) with unequally distributed
levels. margin = "marginalmeans" will “average” over the
levels of non-focal factors, while margin = "empirical"
will average over the observations in your sample.
Let’s show this with a very simple example:
data(iris)
set.seed(123)
# create an unequal distributed factor, used as non-focal term
iris$x <- as.factor(sample(1:4, nrow(iris), replace = TRUE, prob = c(0.1, 0.2, 0.3, 0.4)))
m <- lm(Sepal.Width ~ Species + x, data = iris)
# predicted values, conditioned on x = 1
predict_response(m, "Species")
#> # Predicted values of Sepal.Width
#>
#> Species | Predicted | 95% CI
#> -----------------------------------
#> setosa | 3.50 | 3.30, 3.69
#> versicolor | 2.84 | 2.63, 3.04
#> virginica | 3.04 | 2.85, 3.23
#>
#> Adjusted for:
#> * x = 1
# predicted values, conditioned on weighted average of x
predict_response(m, "Species", margin = "marginalmeans")
#> # Predicted values of Sepal.Width
#>
#> Species | Predicted | 95% CI
#> -----------------------------------
#> setosa | 3.45 | 3.35, 3.54
#> versicolor | 2.78 | 2.68, 2.89
#> virginica | 2.99 | 2.89, 3.09
# average predicted values, averaged over the sample and aggregated by "Species"
predict_response(m, "Species", margin = "empirical")
#> # Average predicted values of Sepal.Width
#>
#> Species | Predicted | 95% CI
#> -----------------------------------
#> setosa | 3.43 | 3.34, 3.52
#> versicolor | 2.77 | 2.67, 2.86
#> virginica | 2.97 | 2.88, 3.07Finally, the weighting for margin = "marginalmeans" can
be changed using the weights argument (for details, see
?emmeans::emmeans), which returns results that are more
similar to margin = "empirical".
# the default is an equally weighted average; "proportional" weights in
# proportion to the frequencies of factor combinations
predict_response(m, "Species", margin = "marginalmeans", weights = "proportional")
#> # Predicted values of Sepal.Width
#>
#> Species | Predicted | 95% CI
#> -----------------------------------
#> setosa | 3.43 | 3.34, 3.52
#> versicolor | 2.77 | 2.67, 2.86
#> virginica | 2.97 | 2.88, 3.07