
Different Backends For Pairwise Comparisons
Source:vignettes/introduction_comparisons_5.Rmd
introduction_comparisons_5.RmdThis vignette is roughly a duplication of the first vignette about Contrasts
and Pairwise Comparisons, but demonstrating the different
backends for the calculation of pairwise comparisons. The default
backend is the marginaleffects
package. If desired, engine = "emmeans" can be used to
switch to the emmeans
package. engine = "ggeffects" is currently experimental and
work-in-progress. It is less feature-rich than the other backends, but
it also works for predictions of random effects.
 Summary of most important points:
Summary of most important points:
- The ggeffects-package relies on the marginaleffects-package by default to calculate contrasts and pairwise comparisons.
-
Although this covers many different ways to test contrasts and comparisons, sometimes it can be convenient or necessary to calculate specific contrasts, like consecutive, interaction or custom contrasts. In this case (e.g. when
test="consecutive", or a data frame for custom contrasts),test_predictions()automatically switches the backend to the emmeans-package . -
The backend can also be changed explicitly by using the
engineargument. Usually, this is not necessary, unless you want to calculate contrasts of random effects levels. This is currently only possible forengine="ggeffects"(see vignette "Case Study: Intersectionality Analysis Using The MAIHDA Framework").
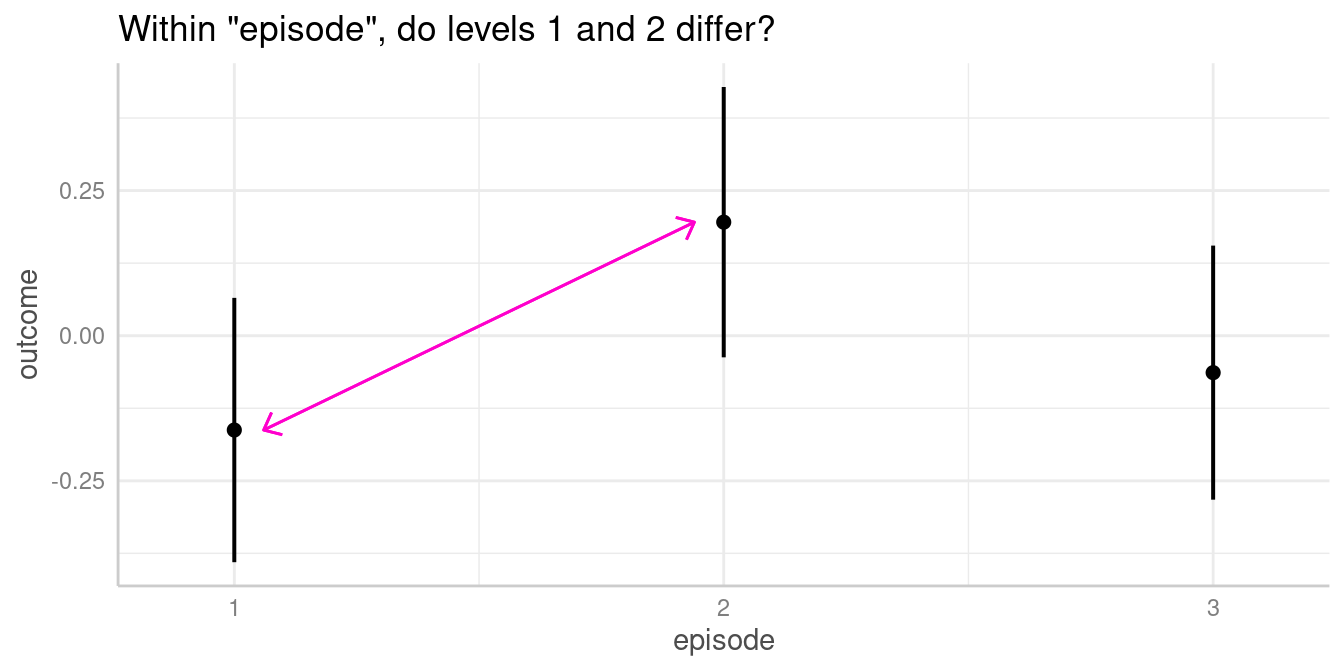
Within episode, do levels differ?
library(ggeffects)
library(ggplot2)
set.seed(123)
n <- 200
d <- data.frame(
outcome = rnorm(n),
grp = as.factor(sample(c("treatment", "control"), n, TRUE)),
episode = as.factor(sample(1:3, n, TRUE)),
sex = as.factor(sample(c("female", "male"), n, TRUE, prob = c(0.4, 0.6)))
)
model1 <- lm(outcome ~ grp + episode + grp, data = d)Predictions
my_pred <- predict_response(model1, "episode", margin = "marginalmeans")
my_pred
#> # Predicted values of outcome
#>
#> episode | Predicted | 95% CI
#> ---------------------------------
#> 1 | -0.16 | -0.39, 0.07
#> 2 | 0.20 | -0.04, 0.43
#> 3 | -0.06 | -0.28, 0.16Pairwise comparisons
# comparisons based on estimated marginal means, using "marginaleffects" package
test_predictions(model1, "episode", margin = "marginalmeans")
#> # Pairwise comparisons
#>
#> episode | Contrast | 95% CI | p
#> -----------------------------------------
#> 1-2 | -0.36 | -0.68, -0.03 | 0.031
#> 1-3 | -0.10 | -0.42, 0.22 | 0.538
#> 2-3 | 0.26 | -0.06, 0.58 | 0.112
# comparisons using "emmeans" package
test_predictions(model1, "episode", engine = "emmeans")
#> # Pairwise comparisons
#>
#> episode | Contrast | 95% CI | p
#> -----------------------------------------
#> 1-2 | -0.36 | -0.68, -0.03 | 0.031
#> 1-3 | -0.10 | -0.42, 0.22 | 0.538
#> 2-3 | 0.26 | -0.06, 0.58 | 0.112
# comparisons using "ggeffects" backend. This engine requires the
# ggeffects-object as input
test_predictions(my_pred, engine = "ggeffects")
#> # Pairwise comparisons
#>
#> episode | Contrast | 95% CI | p
#> -----------------------------------------
#> 1-2 | -0.36 | -0.68, -0.03 | 0.031
#> 1-3 | -0.10 | -0.41, 0.22 | 0.538
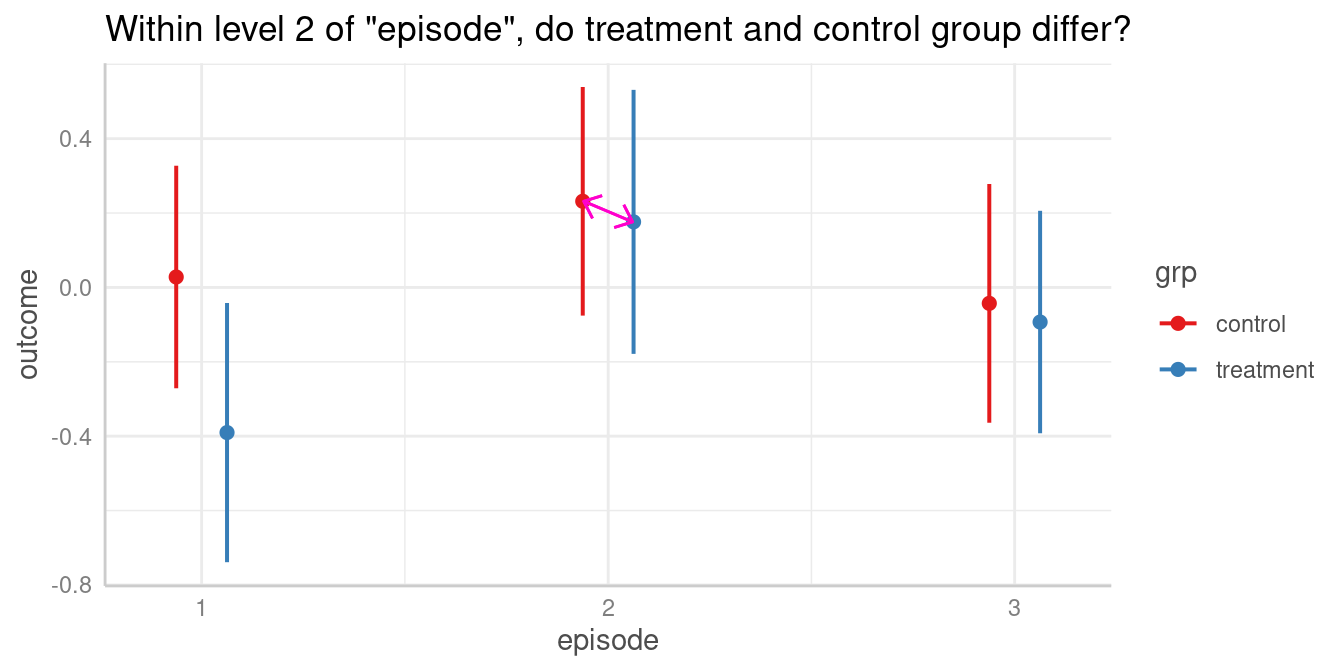
#> 2-3 | 0.26 | -0.06, 0.58 | 0.111Does same level of episode differ between groups?
model2 <- lm(outcome ~ grp * episode + grp, data = d)Predictions
my_pred <- predict_response(model2, c("episode", "grp"), margin = "marginalmeans")
my_pred
#> # Predicted values of outcome
#>
#> grp: control
#>
#> episode | Predicted | 95% CI
#> ----------------------------------
#> 1 | 0.03 | -0.27, 0.33
#> 2 | 0.23 | -0.08, 0.54
#> 3 | -0.04 | -0.36, 0.28
#>
#> grp: treatment
#>
#> episode | Predicted | 95% CI
#> ----------------------------------
#> 1 | -0.39 | -0.74, -0.04
#> 2 | 0.18 | -0.18, 0.53
#> 3 | -0.09 | -0.39, 0.21Pairwise comparisons
# we want "episode = 2-2" and "grp = control-treatment"
# comparisons based on estimated marginal means, using "marginaleffects" package
test_predictions(model2, c("episode [2]", "grp"), margin = "marginalmeans")
#> # Pairwise comparisons
#>
#> grp | Contrast | 95% CI | p
#> --------------------------------------------------
#> control-treatment | 0.06 | -0.41, 0.52 | 0.816
# comparisons based using "emmeans" package
test_predictions(model2, c("episode [2]", "grp"), engine = "emmeans")
#> # Pairwise comparisons
#>
#> episode | grp | Contrast | 95% CI | p
#> ------------------------------------------------------------
#> 2-2 | control-treatment | 0.06 | -0.41, 0.52 | 0.816
# comparisons using "ggeffects" backend
my_pred <- predict_response(model2, c("episode [2]", "grp"), margin = "marginalmeans")
test_predictions(my_pred, engine = "ggeffects")
#> # Pairwise comparisons
#>
#> episode | grp | Contrast | 95% CI | p
#> ------------------------------------------------------------
#> 2-2 | control-treatment | 0.06 | -0.41, 0.52 | 0.816Does difference between two levels of episode in the control group differ from difference of same two levels in the treatment group?
The test argument also allows us to compare
difference-in-differences. When engine = "emmeans" or
"ggeffects", we need to set
test = "interaction" to get interaction contrasts,
i.e. differences-in-differences.
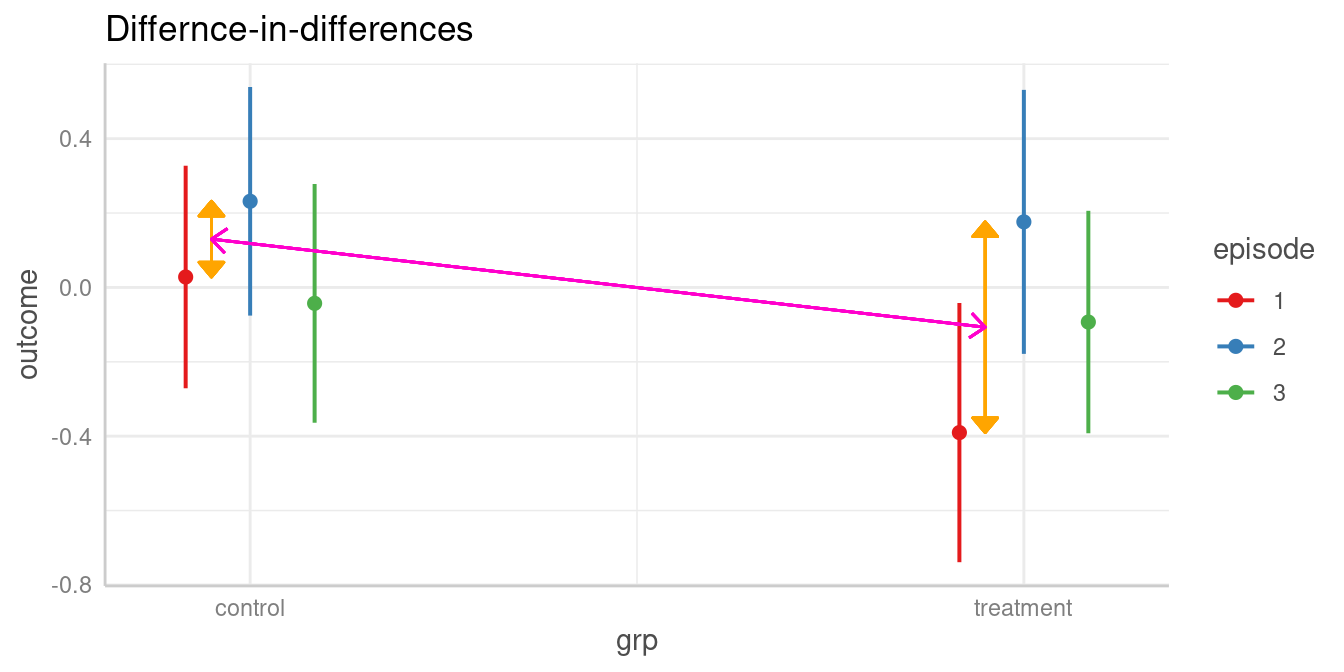
# specifying the difference-in-difference when using "marginaleffects"
test_predictions(model2, c("episode", "grp"), test = "(b1 - b3) = (b2
- b4)", margin = "marginalmeans")
#> Hypothesis | Contrast | 95% CI | p
#> ------------------------------------------------
#> (b1-b3)=(b2-b4) | 0.36 | -0.29, 1.02 | 0.277
#>
#> Tested hypothesis: (episode[1],grp[control] - episode[2],grp[control]) = (episode[1],grp[treatment] - episode[2],grp[treatment])
# "emmeans" provides similar comparisons when we set test = "interaction".
# This displays *all* possible differences-in-differences. The first row in
# this output is identical to the above result from "marginaleffects". The
# "emmeans" package is used automatically, when test = "interaction".
test_predictions(model2, c("episode", "grp"), test = "interaction")
#> # Interaction contrasts
#>
#> episode | grp | Contrast | 95% CI | p
#> ----------------------------------------------------------------
#> 1-2 | control and treatment | 0.36 | -0.29, 1.02 | 0.277
#> 1-3 | control and treatment | 0.37 | -0.27, 1.00 | 0.254
#> 2-3 | control and treatment | 0.01 | -0.64, 0.65 | 0.988
# using "ggeffects", we also need to set test = "interaction" to get the same
# results. However, since by default "emmeans" us used, we also need to specify
# the "engine" argument
my_pred <- predict_response(model2, c("episode", "grp"), margin = "marginalmeans")
test_predictions(my_pred, test = "interaction", engine = "ggeffects")
#> # Interaction contrasts
#>
#> episode | grp | Contrast | 95% CI | p
#> ----------------------------------------------------------------
#> 1-2 | control and treatment | 0.36 | -0.29, 1.02 | 0.277
#> 1-3 | control and treatment | 0.37 | -0.27, 1.00 | 0.254
#> 2-3 | control and treatment | 0.01 | -0.64, 0.65 | 0.988